Homologous recombination
(HR) takes place during DNA replication at arrested replication forks, where it promotes the stability of the fork itself and, when the fork is inactivated, promotes the reconstitution of replication. HR also participates in the repair of double strand breaks (DSBs) in interphase and is essential for meiosis, where sister chromosome cohesion and genetic diversity are generated through programmed DSBs and their subsequent repair. HR is thus essential for the maintenance of genome stability, but since inappropriate recombination can lead to deleterious effects such as loss of heterozygosity and chromosome rearrangements, resulting in cancer in higher organisms, it must be tightly regulated because. This role is fullfilled by homologous recombination mediators which are the focus of our research.
Rrp1-Rrp2 is a new homologous recombination mediator complex identified by us in Schizosaccharomyces pombe, acting together with Rad51 recombinase in a pathway dependent on Swi5-Sfr1 complex and probably negatively regulating one or more sub-pathways of Rad51-mediated recombination together with antirecombinogenic helicase Srs2. Rrp1 and Rrp2 are paralogues of Saccharomyces cerevisiae Uls1 involved, as we have demonstrated, in the replication stress response.
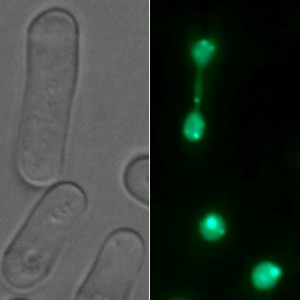
Rrp1 and Rrp2 bind to DNA at the sites of double strand breaks what can be observed under fluorescent microscope as colocalising foci in the nuclei of the cells. Now, using fluorescence microscope obrervations and coimmunoprecipitation we examine the interactions of Rrp1 and Rrp2 with other proteins involved in the maintenance of genome stability. We also test the role the 3 motifs present in Rrp1 and Rrp2 sequences – SNF2-N (characteristic for chromatin remodelers) and Helicase C (present in DNA dependent ATPases) and zf-C3HC4, zinc finger (suggesting a role in protein ubiquitination) may have for these proteins activity.
Principal investigator: dr Dorota Dziadkowiec
Cooperation:
1. Prof. Robert Wysocki – Department of Genetics and Cell Physiology, Faculty of Biological Sciences, Wrocław University
2. Prof. Antony M. Carr – Genome Damage and Stability Center, University of Sussex, United Kingdom
3. Prof. Hiroshi Iwasaki – Tokyo Institute of Technology, Japan
PhD students:
1. Karol Kramarz – The role of Rad5 ortologues in DNA repair in Saccharomyces cerevisiae and Schizosaccharomyces pombe.
2. Anna Barg – The role of homologous recombination mediators in the maintenance of centromere integrity.
3. Jakub Muraszko – The role of homologous recombination mediators in the maintenance of chromatin structure.
List of publications:
- K. Kramarz, I. Litwin, M. Cal-Bąkowska, B. Szakal, D. Branzei, R. Wysocki, D. Dziadkowiec (2014) Swi2/Snf2-like protein Uls1 functions in the Sgs1-dependent pathway of maintenance of rDNA stability and alleviation of replication stress. DNA Repair 21:24-35
- D. Dziadkowiec, K. Kramarz, K. Kanik, P. Wisniewski, A.M. Carr (2013) Involvement of Schizosaccharomyces pombe rrp1+ and rrp2+ in the Srs2- and Swi5/Sfr1-dependent pathway in response to DNA damage and replication inhibition. Nucl Acids Res. 41:8196-8209.
- I. Litwin, T. Bocer, D. Dziadkowiec, R. Wysocki (2013) Oxidative stress and replication-independent DNA breakage induced by arsenic in Saccharomyces cerevisiae. PLoS Genet. 9:e1003640.
- M. Cal-Bąkowska, I. Litwin, T. Bocer, R. Wysocki, D. Dziadkowiec (2011) The Swi2–Snf2-like protein Uls1 is involved in replication stress response. Nuclc Acids Res 39: 8765–8777
- D. Dziadkowiec, E. Petters, A. Dyjankiewicz, P. Karpiński, V. Garcia, A. Watson, A.M. Carr (2009) The role of novel genes rrp1+ and rrp2+ in the repair of DNA damage in Schizosaccharomyces pombe. DNA Repair 8: 627-636









